Welcome to the ModelBricks project
|
|
Many computational models in biology are built and intended for "single-use"; the modeling assumptions and
descriptions of model elements must be found in the corresponding paper. Expanding models to new and more complex situations is non-trivial.
As a result, new models are almost always created anew, repeating literature searches for kinetic parameters, initial conditions and modeling specifics. It is akin to building a brick house starting with a pile of clay. Here we present a database of ModelBricks - well-annotated, reusable component mechanisms that can be assembled into
fully annotated mathematical models for cell biology.
Browse ModelBricks |
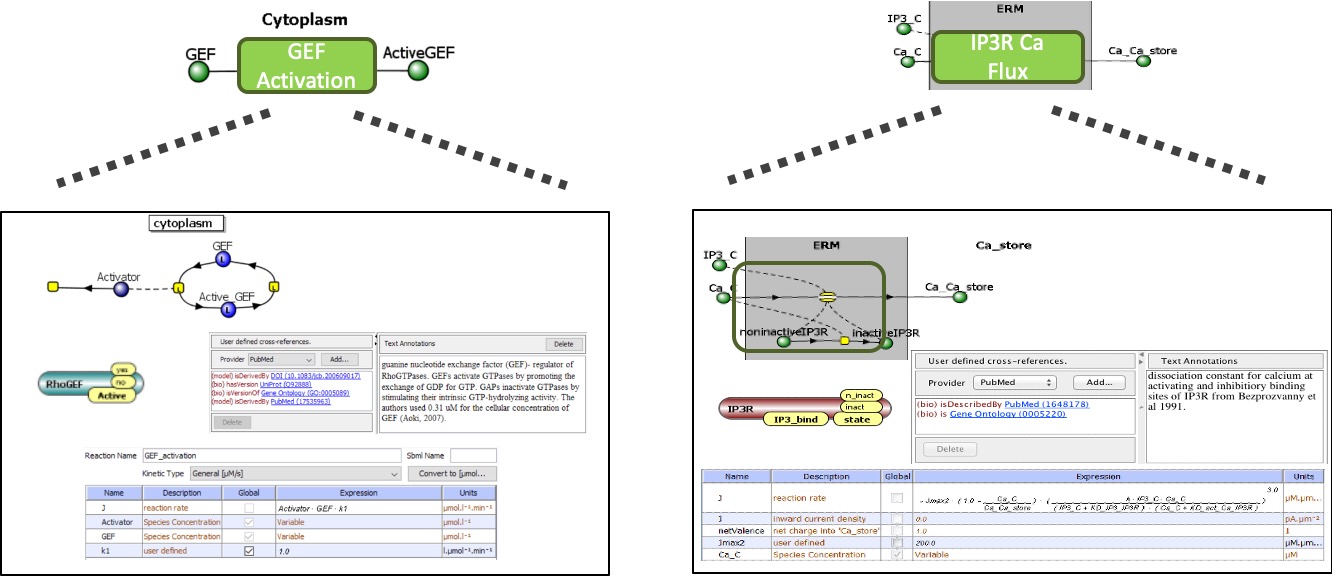
Each ModelBrick contains
|
1. Sets of molecules (species) uniquely identified with universal identifiers, their initial concentrations and transport properties. |
|
2. Reaction(s) (changes in state) identified by known biochemical mechanisms and defined by a flux reaction with parameters for forward and reverse rates (kf and kr). Flux terms can be simple mass action kinetics or complex functions. |
|
3. An identified cellular Compartment(s) where the reaction occurs and the species reside. |
|
4. A Publication ID or other identifier of the author and peer-review status for the original model from which a ModelBrick is derived |
ModelBricks can be
|
1. Downloaded in mulltiple formats, such as SBML or VCML formats |
|
2. Inserted into an existing SBML compatible model including all annotations and defined parameters. |
|
3.Created from existing models and submitted for inclusion in the repository. |
Browse ModelBricks
